Number Preprocessing#
import os
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
%matplotlib inline
data_path = os.path.expanduser("../../data/diabetes.csv")
df = pd.read_csv(data_path)
df.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
Inspecting the dataset#
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 768 entries, 0 to 767
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Pregnancies 768 non-null int64
1 Glucose 768 non-null int64
2 BloodPressure 768 non-null int64
3 SkinThickness 768 non-null int64
4 Insulin 768 non-null int64
5 BMI 768 non-null float64
6 DiabetesPedigreeFunction 768 non-null float64
7 Age 768 non-null int64
8 Outcome 768 non-null int64
dtypes: float64(2), int64(7)
memory usage: 54.1 KB
df.describe()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| count | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 |
| mean | 3.845052 | 120.894531 | 69.105469 | 20.536458 | 79.799479 | 31.992578 | 0.471876 | 33.240885 | 0.348958 |
| std | 3.369578 | 31.972618 | 19.355807 | 15.952218 | 115.244002 | 7.884160 | 0.331329 | 11.760232 | 0.476951 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.078000 | 21.000000 | 0.000000 |
| 25% | 1.000000 | 99.000000 | 62.000000 | 0.000000 | 0.000000 | 27.300000 | 0.243750 | 24.000000 | 0.000000 |
| 50% | 3.000000 | 117.000000 | 72.000000 | 23.000000 | 30.500000 | 32.000000 | 0.372500 | 29.000000 | 0.000000 |
| 75% | 6.000000 | 140.250000 | 80.000000 | 32.000000 | 127.250000 | 36.600000 | 0.626250 | 41.000000 | 1.000000 |
| max | 17.000000 | 199.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 | 1.000000 |
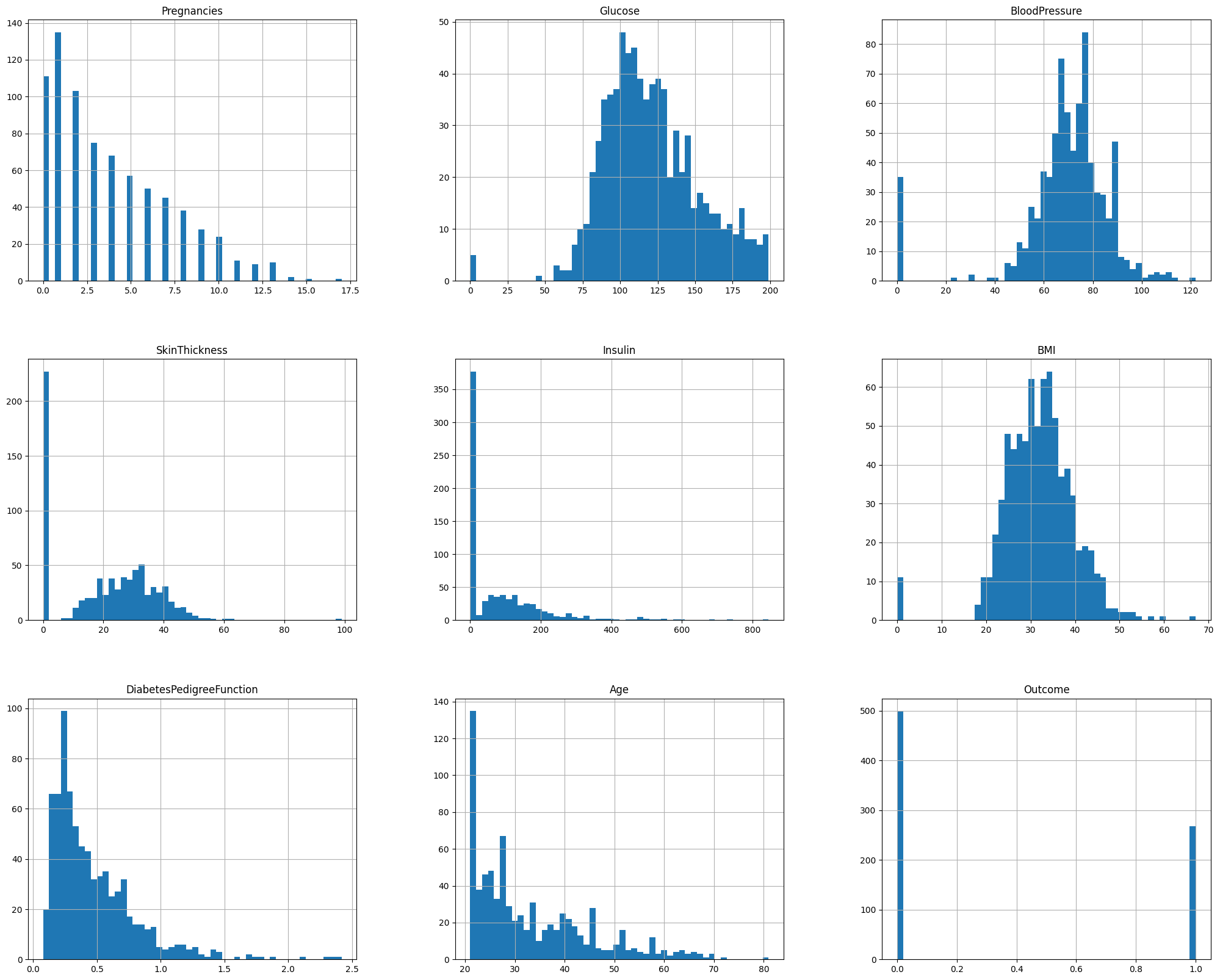
Visualizing the dataset#
df.hist(bins=50, figsize=(25, 20))
plt.show()
Removing duplicated data points#
df.drop_duplicates(keep="first", inplace=True)
Creating training and testing set#
train_df, test_df = train_test_split(df, test_size=0.2)
Gaining further insight#
correlation_matrix = train_df.corr(method="pearson")
correlation_matrix
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| Pregnancies | 1.000000 | 0.135109 | 0.164122 | -0.041149 | -0.056988 | 0.017513 | -0.015260 | 0.537494 | 0.217668 |
| Glucose | 0.135109 | 1.000000 | 0.164170 | 0.048504 | 0.328315 | 0.210757 | 0.125436 | 0.269642 | 0.475968 |
| BloodPressure | 0.164122 | 0.164170 | 1.000000 | 0.214924 | 0.080210 | 0.249102 | 0.044764 | 0.241728 | 0.114867 |
| SkinThickness | -0.041149 | 0.048504 | 0.214924 | 1.000000 | 0.424873 | 0.391442 | 0.176407 | -0.088288 | 0.097148 |
| Insulin | -0.056988 | 0.328315 | 0.080210 | 0.424873 | 1.000000 | 0.193953 | 0.140380 | -0.034457 | 0.140022 |
| BMI | 0.017513 | 0.210757 | 0.249102 | 0.391442 | 0.193953 | 1.000000 | 0.126622 | 0.020420 | 0.306521 |
| DiabetesPedigreeFunction | -0.015260 | 0.125436 | 0.044764 | 0.176407 | 0.140380 | 0.126622 | 1.000000 | 0.051460 | 0.170353 |
| Age | 0.537494 | 0.269642 | 0.241728 | -0.088288 | -0.034457 | 0.020420 | 0.051460 | 1.000000 | 0.234581 |
| Outcome | 0.217668 | 0.475968 | 0.114867 | 0.097148 | 0.140022 | 0.306521 | 0.170353 | 0.234581 | 1.000000 |
correlation_matrix["Outcome"].sort_values()
SkinThickness 0.097148
BloodPressure 0.114867
Insulin 0.140022
DiabetesPedigreeFunction 0.170353
Pregnancies 0.217668
Age 0.234581
BMI 0.306521
Glucose 0.475968
Outcome 1.000000
Name: Outcome, dtype: float64
Handling missing data#
names = [
"Glucose",
"BloodPressure",
"SkinThickness",
"Insulin",
"BMI",
"DiabetesPedigreeFunction",
"Age",
]
for name in names:
train_df[name].replace(0, np.nan, inplace=True)
/var/folders/7w/fv5n0x414253d7dv5g2wwmb40000gn/T/ipykernel_1701/551462321.py:4: FutureWarning: A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
train_df[name].replace(0, np.nan, inplace=True)
train_df.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 683 | 4 | 125.0 | 80.0 | NaN | NaN | 32.3 | 0.536 | 27 | 1 |
| 394 | 4 | 158.0 | 78.0 | NaN | NaN | 32.9 | 0.803 | 31 | 1 |
| 190 | 3 | 111.0 | 62.0 | NaN | NaN | 22.6 | 0.142 | 21 | 0 |
| 274 | 13 | 106.0 | 70.0 | NaN | NaN | 34.2 | 0.251 | 52 | 0 |
| 162 | 0 | 114.0 | 80.0 | 34.0 | 285.0 | 44.2 | 0.167 | 27 | 0 |
glucose_median = train_df["Glucose"].median()
blood_pressure_median = train_df["BloodPressure"].median()
skin_thickness_median = train_df["SkinThickness"].median()
insulin_median = train_df["Insulin"].median()
bmi_median = train_df["BMI"].median()
age_median = train_df["Age"].median()
dpf_median = train_df["DiabetesPedigreeFunction"].median()
train_df["Glucose"].fillna(glucose_median, inplace=True)
train_df["BloodPressure"].fillna(blood_pressure_median, inplace=True)
train_df["SkinThickness"].fillna(skin_thickness_median, inplace=True)
train_df["Insulin"].fillna(insulin_median, inplace=True)
train_df["BMI"].fillna(bmi_median, inplace=True)
train_df["Age"].fillna(age_median, inplace=True)
train_df["DiabetesPedigreeFunction"].fillna(dpf_median, inplace=True)
/var/folders/7w/fv5n0x414253d7dv5g2wwmb40000gn/T/ipykernel_1701/2039840509.py:13: FutureWarning: A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
train_df["BMI"].fillna(bmi_median, inplace=True)
/var/folders/7w/fv5n0x414253d7dv5g2wwmb40000gn/T/ipykernel_1701/2039840509.py:14: FutureWarning: A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
train_df["Age"].fillna(age_median, inplace=True)
/var/folders/7w/fv5n0x414253d7dv5g2wwmb40000gn/T/ipykernel_1701/2039840509.py:15: FutureWarning: A value is trying to be set on a copy of a DataFrame or Series through chained assignment using an inplace method.
The behavior will change in pandas 3.0. This inplace method will never work because the intermediate object on which we are setting values always behaves as a copy.
For example, when doing 'df[col].method(value, inplace=True)', try using 'df.method({col: value}, inplace=True)' or df[col] = df[col].method(value) instead, to perform the operation inplace on the original object.
train_df["DiabetesPedigreeFunction"].fillna(dpf_median, inplace=True)
train_df.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 683 | 4 | 125.0 | 80.0 | 29.0 | 125.0 | 32.3 | 0.536 | 27 | 1 |
| 394 | 4 | 158.0 | 78.0 | 29.0 | 125.0 | 32.9 | 0.803 | 31 | 1 |
| 190 | 3 | 111.0 | 62.0 | 29.0 | 125.0 | 22.6 | 0.142 | 21 | 0 |
| 274 | 13 | 106.0 | 70.0 | 29.0 | 125.0 | 34.2 | 0.251 | 52 | 0 |
| 162 | 0 | 114.0 | 80.0 | 34.0 | 285.0 | 44.2 | 0.167 | 27 | 0 |
Encoding categorial attributes#
# Copy original dataframe and add new column with random fitness values
temp_df = train_df.copy()
fitness_values = [
"bad",
"moderate",
"good",
"very good",
]
temp_df["fitness"] = np.random.choice(fitness_values, temp_df.shape[0])
temp_df.head(5)
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | fitness | |
|---|---|---|---|---|---|---|---|---|---|---|
| 683 | 4 | 125.0 | 80.0 | 29.0 | 125.0 | 32.3 | 0.536 | 27 | 1 | very good |
| 394 | 4 | 158.0 | 78.0 | 29.0 | 125.0 | 32.9 | 0.803 | 31 | 1 | very good |
| 190 | 3 | 111.0 | 62.0 | 29.0 | 125.0 | 22.6 | 0.142 | 21 | 0 | bad |
| 274 | 13 | 106.0 | 70.0 | 29.0 | 125.0 | 34.2 | 0.251 | 52 | 0 | moderate |
| 162 | 0 | 114.0 | 80.0 | 34.0 | 285.0 | 44.2 | 0.167 | 27 | 0 | very good |
from sklearn.preprocessing import LabelEncoder
encoder = LabelEncoder()
fitness_encoded = encoder.fit_transform(temp_df["fitness"])
for id_, class_ in enumerate(encoder.classes_):
print(f"class id {id_} has label {class_}")
print()
print(f"Encoded fitness values for first 10 entries: {fitness_encoded[:10]}")
class id 0 has label bad
class id 1 has label good
class id 2 has label moderate
class id 3 has label very good
Encoded fitness values for first 10 entries: [3 3 0 2 3 0 1 3 2 1]
Rescaling or standardizing attributes#
from sklearn.preprocessing import MinMaxScaler
# initialize min-max scaler
mm_scaler = MinMaxScaler()
temp1_df = train_df.copy()
column_names = temp1_df.columns.tolist()
# transform all attributes
temp1_df[column_names] = mm_scaler.fit_transform(temp1_df[column_names])
temp1_df.sort_index(inplace=True)
temp1_df.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 0.470588 | 0.896774 | 0.444444 | 0.239130 | 0.133413 | 0.104294 | 0.253629 | 0.183333 | 1.0 |
| 3 | 0.058824 | 0.290323 | 0.466667 | 0.173913 | 0.096154 | 0.202454 | 0.038002 | 0.000000 | 0.0 |
| 5 | 0.294118 | 0.464516 | 0.555556 | 0.239130 | 0.133413 | 0.151329 | 0.052519 | 0.150000 | 0.0 |
| 6 | 0.176471 | 0.219355 | 0.288889 | 0.271739 | 0.088942 | 0.261759 | 0.072588 | 0.083333 | 1.0 |
| 7 | 0.588235 | 0.458065 | 0.533333 | 0.239130 | 0.133413 | 0.349693 | 0.023911 | 0.133333 | 0.0 |
from sklearn.preprocessing import StandardScaler
standard_scaler = StandardScaler()
temp2_df = train_df.copy()
# transform all attributes
temp2_df[column_names] = mm_scaler.fit_transform(temp2_df[column_names])
temp2_df.sort_index(inplace=True)
temp2_df.head()
| Pregnancies | Glucose | BloodPressure | SkinThickness | Insulin | BMI | DiabetesPedigreeFunction | Age | Outcome | |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 0.470588 | 0.896774 | 0.444444 | 0.239130 | 0.133413 | 0.104294 | 0.253629 | 0.183333 | 1.0 |
| 3 | 0.058824 | 0.290323 | 0.466667 | 0.173913 | 0.096154 | 0.202454 | 0.038002 | 0.000000 | 0.0 |
| 5 | 0.294118 | 0.464516 | 0.555556 | 0.239130 | 0.133413 | 0.151329 | 0.052519 | 0.150000 | 0.0 |
| 6 | 0.176471 | 0.219355 | 0.288889 | 0.271739 | 0.088942 | 0.261759 | 0.072588 | 0.083333 | 1.0 |
| 7 | 0.588235 | 0.458065 | 0.533333 | 0.239130 | 0.133413 | 0.349693 | 0.023911 | 0.133333 | 0.0 |

